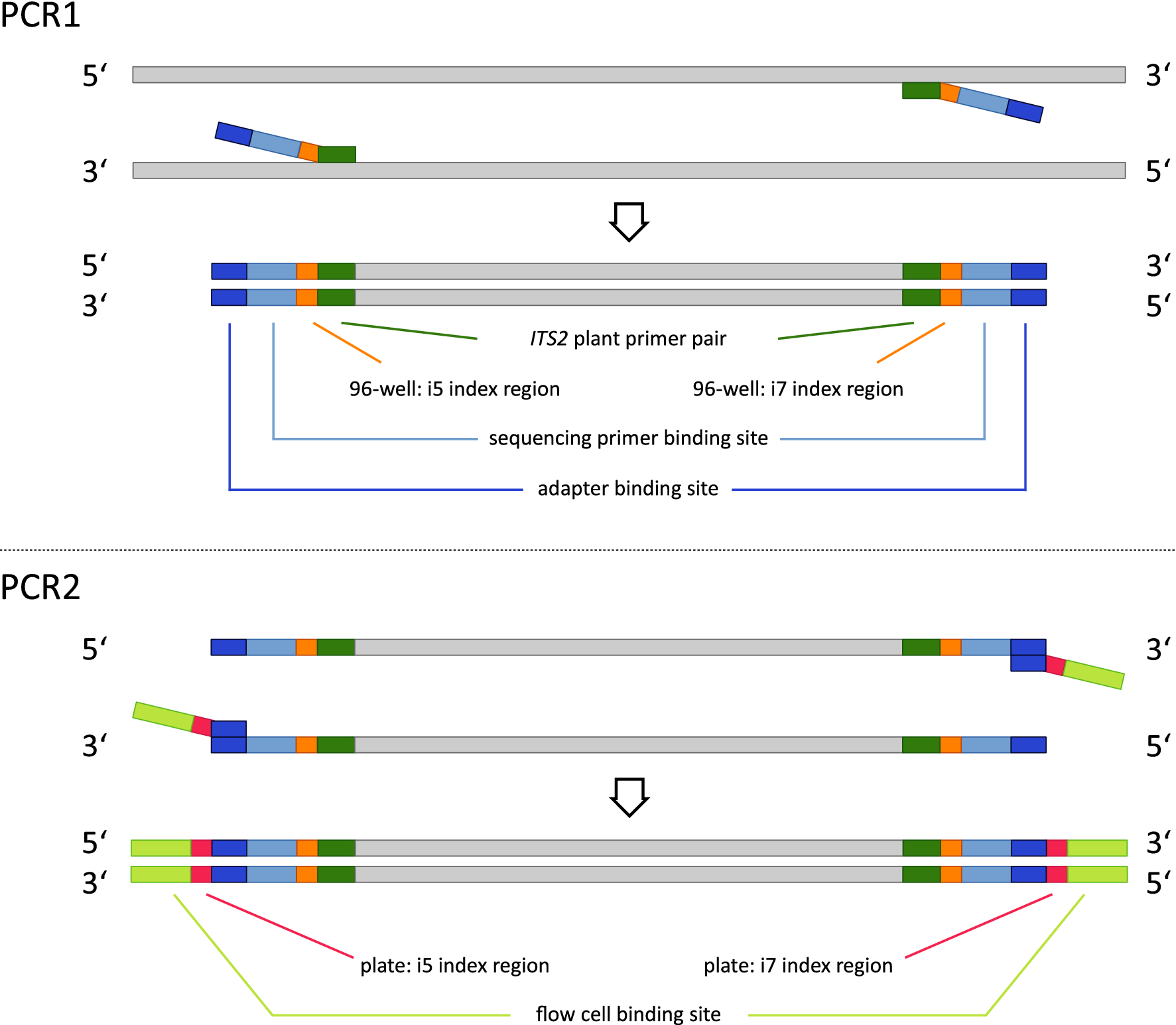
Large Scale Loss of Data in Low-Diversity Illumina Sequencing Libraries Can Be Recovered by Deferred Cluster Calling | PLOS ONE

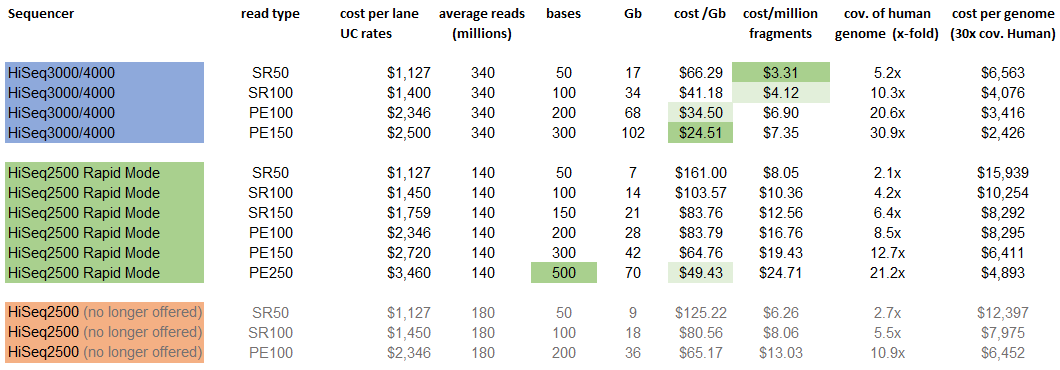
Increased read duplication on patterned flowcells- understanding the impact of Exclusion Amplification - Enseqlopedia
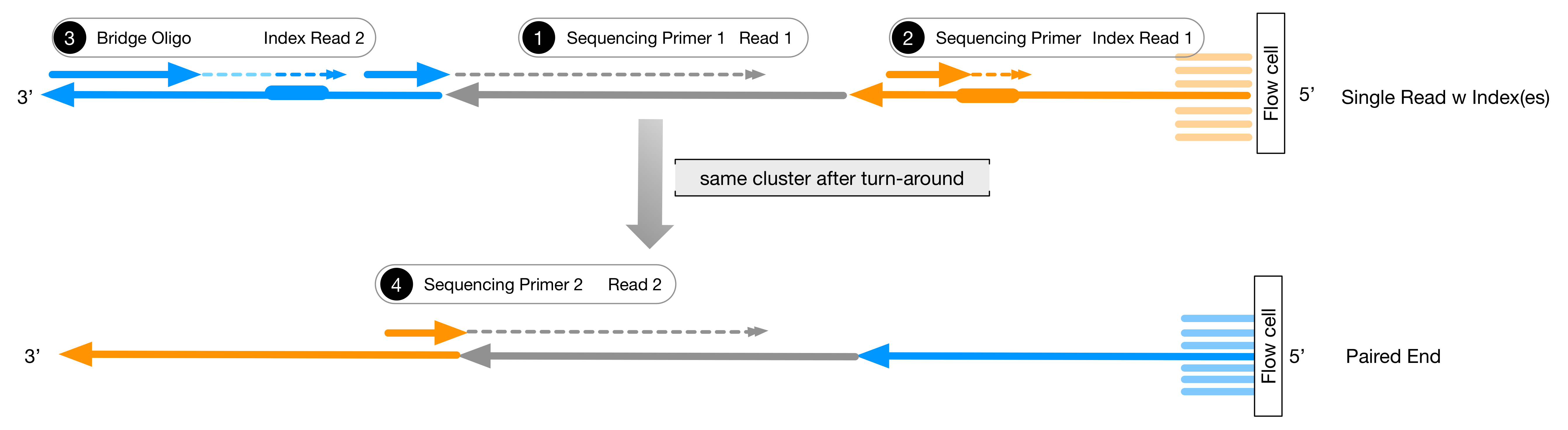
Illumina Sequencing (for Dummies) -An overview on how our samples are sequenced. – kscbioinformatics

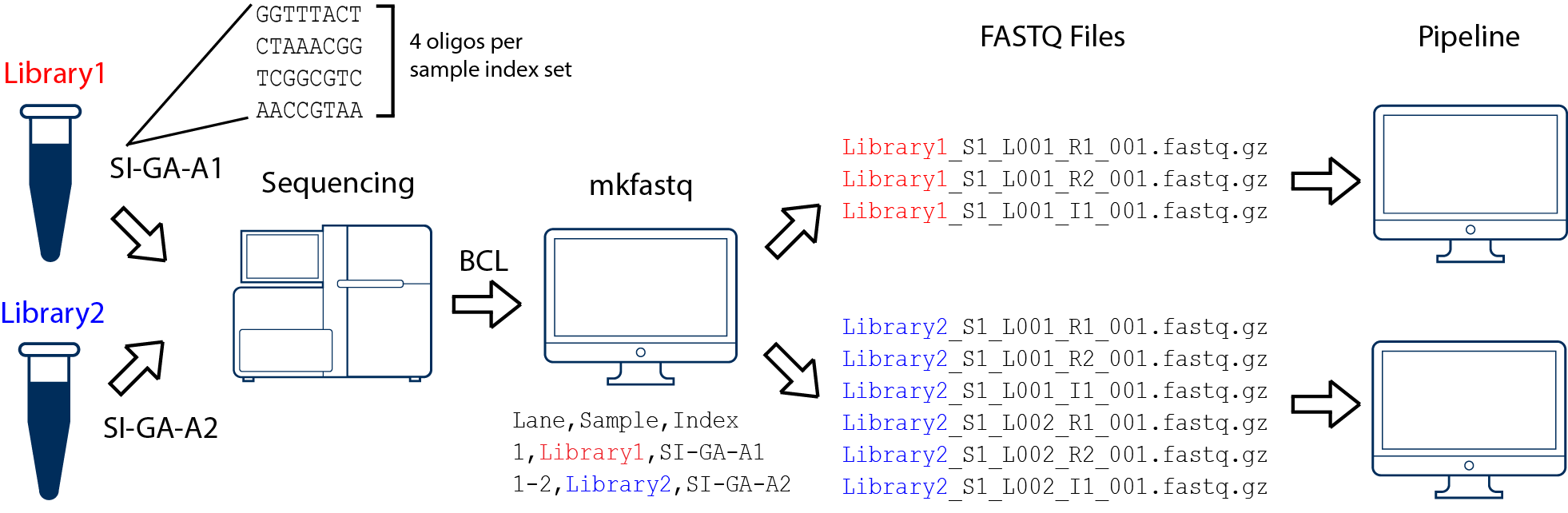
An open source toolkit for repurposing Illumina sequencing systems as versatile fluidics and imaging platforms | Scientific Reports